How do bacteria regulate their metabolism?
A global control mechanism adapts bacteria to changing environments
2017-11-29 – News from the Physics Department

On earth parameters like temperature, light conditions, availability of food and many other essential aspects of the environment fluctuate in time and vary from place to place. Life as we know it is only possible, because nature has the means to deal with many types of such environmental fluctuations. Each cell and every organism relies on countless mechanisms to adapt.
Professor Ulrich Gerland explains: “This is especially true for bacteria, which cannot shape their environment or migrate large distances. They have to more or less cope with their environment as it is.”
One of the best studied model systems is Escherichia coli, a bacterium which lives in the gut of mammals, including humans. It has to adapt to the constantly changing nutrients that flow by, and produce specific enzymes only when needed. For instance, the enzymes that break down the milk sugar lactose are only needed when lactose is actually present; other enzymes that help synthesize, e.g., certain amino acids, are only required when these amino acids are missing.
Escherichia coli adapt to different food supplies
In the lab scientists study these adaptations of the microorganisms by changing the nutrition of bacteria – i.e. the composition of the brew in which they nourish. It is well established, and earned Jacques Monod a Nobel Prize in 1965, that bacteria adapt to the current environment by regulating the expression (and thus the number of copies) of particular proteins. Thus, depending on environmental factors, the concentrations of different proteins are adjusted.
However, despite the great interest and the tremendous research carried out over half a century, the biochemical details of this complicated mechanism from the bacterium sensing what is required to the actual regulation of the concentration of enzymes are still far from fully understood. This is not surprising, if one keeps in mind, that even a rather “simple” bacterium as E. coli has several thousand different kinds of proteins and other molecules (in different concentrations) packed tightly within its tiny microscopic enclosure and a corresponding DNA containing thousands of genes.
“At present it is impossible to account for all the interactions and biochemical reactions between all molecules present in a bacterium,” says Ulrich Gerland. “All the biomolecules present and all reactions that take place within the cell are part of the bacterium’s metabolism.” Bacterial growth by cell division essentially requires that the full set of biomolecules within a cell is duplicated from scratch.
Growth transition kinetics
His team together with colleagues from the University of California at San Diego has taken an approach that focuses on the principal mechanisms of these regulation processes instead of looking at the molecular details of the reaction pathways. The fundamental question the team of researchers addressed experimentally is on how fast do bacteria adapt to different changes of the environment?
They performed a large set of experiments where the growth conditions for the bacteria were changed. For example, by adding a good nutrient source after a period of poor supply of food or vice versa. At so-called up-shifts of nutrients, the growth rate of the bacteria increases after some lag due to the adaption process.
Other experiments induced a so-called diauxie, a phenomenon already observed by Monod. If bacteria are fed on one type of food and later on a different kind, growth slows or comes to a halt, although there is always enough food present. This is due to the bacteria shifting their digestion - i.e. the concentrations of certain enzymes - from one food to another.
By detailed observations of different parameters during the development of the bacterial cultures a quantitative characterization of the temporal evolution of the regulation was achieved. As one would expect, the different experiments yield quantitatively and qualitatively different growth curves. The measured transition curves are specific to the respective start parameters and to the shifts induced.
Model of flux-controlled regulation (FCR)
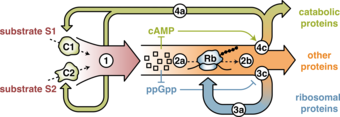
The scientists did not stop at these observations. In order to elucidate the bacteria’s adaption strategy, they developed a model to describe the phenomena from a physicist’s perspective. In a top-down approach, the model just uses a qualitative knowledge of the biochemical details of the regulation. It is based on balancing the flux of substances within the bacterium, i.e. establishing equations on the flow of matter.
Several regulatory processes are considered, but it turns out that applying the condition of a balanced flux of substances, these can be simplified to just a single differential equation, describing the global kinetics of the whole set of biochemical reactions involved in the different adaptions.
“Our steady-state model of the regulation mechanism correctly describes the temporal development of adaptation to changing nutrients, as well as increases, reductions and changes in the available nutrients, quantitatively and without adjustable parameters,” says Ulrich Gerland, summarizing the results of the study.
“Apparently, the kinetics of growth adaptation do not depend on microscopic details of the individual biochemical reactions, but rather adhere to a global strategy for the redistribution of resources for protein synthesis,” says Ulrich Gerland. “It is thus conceivable that our theoretical model might be applicable to an array of similar kinetic processes.”
The research was funded by the National Institutes of Health (NIH), USA, by the Simons Foundation, USA, and the German Research Foundation (DFG) via the Excellence Cluster ‘Nanosystems Initiative Munich’, as well as the priority program SPP1617.
- Editing
- Dr. Johannes Wiedersich
Publications
- A global resource allocation strategy governs growth transition kinetics of Escherichia coliDavid W. Erickson, Severin J. Schink, Vadim Patsalo, James R. Williamson, Ulrich Gerland & Terence HwaDOI: 10.1038/nature24299
- Quantifying the benefit of a proteome reserve in fluctuating environmentsMatteo Mori, Severin Schink, David W. Erickson, Ulrich Gerland, and Terence Hwa